Proof of life using molecular tools
Researchers from the Cawthron Institute used samples of bilge water (i.e., sea water that accumulates on-board a vessel during its operations) collected from 15 small recreational and commercial vessels in the Nelson and Marlborough regions for the study. The planktonic life stages of non-native marine species can potentially be transported in the bilge water of small vessels, but only represent a biosecurity threat if they are discharged alive into a new environment.
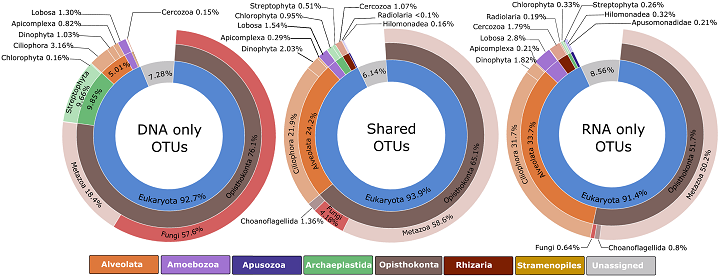
Environmental DNA and RNA molecules were extracted from the bilge water samples and sequenced to identify the organisms that they were associated with. Because RNA molecules degrade much more quickly in natural environments than DNA, they are likely to be better indicators of living ("viable") organisms in the samples.The study compared the numbers of organisms that were identified only from analysis of DNA, only from RNA, and those that were retrieved using both methods.
Just over 62% of the organisms identified from the bilge water were found using both methods. A further 19% were recorded only in the DNA analyses and 17% were unique to the RNA method. For biosecurity applications that require detection of living organisms, the study suggests using only organisms that were present in both eDNA and eRNA analyses.
Further research is needed to improve understanding of the persistence of RNA in the environment, and the underlying reasons for the presence of RNA-only organisms in environmental samples
The research is published in the Open Access journal PlosONE and was funded by NIWA through the Strategic Science Investment Fund [Coasts and Oceans Research Programme 6 (Marine Biosecurity)].
Additional reading
- Pochon, X., Zaiko, A., Fletcher, L.M., Laroche, O., Wood, S.A. (2017) Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications. PLoS ONE, 12(11): e0187636.10.1371/journal.pone.0187636
- Can marine pests be transported and spread in bilge water?
- Fletcher, L., Zaiko, A., Atalah, J., Richter, I., Dufour, C., Pochon, X., Wood, S., Hopkins, G. (2017) Bilge water as a vector for the spread of marine pests: a morphological, metabarcoding and experimental assessment. Biological Invasions, 19: 2851–2867. doi:10.1007/s10530-017-1489-y
Key contacts
| Xavier Pochon |
| [email protected] |
 |